 | Velocity Estimation of Spots in Three-Dimensional Confocal Image Sequences of Living Cells In Cytometry 2001. [bibtex] [pdf] [url] |
Abstract
Background: The analysis of three-dimensional (3D) motion is becoming increasingly important in life cell imaging. A simple description of sometimes complex patterns of movement in living cells gives insight in the underlying mechanisms governing these movements. Methods: We evaluate a velocity estimation method based on intensity derivatives in spatial and temporal domain from 3D confocal images of living cells. Cells of the sample contain intense spots throughout the cell nucleus. In simulations, we model these spots as Gaussian intensity profiles which are constant in intensity and shape. To quantify the quality of the estimated velocity, we introduce a reliability measure. Results: For constant linear velocity, the velocity estimation is unbiased. For accelerated motion paths or when a neighboring spot disturbs the intensity profile, the method results are biased. The influence of the pointspread function on the velocity estimation can be compensated for by introducing anisotropic derivative kernels. The insight gained in the simulations is confirmed by the results of the method applied on an image sequence of a living cell with fluorescently labeled chromatin. Conclusions: With the velocity estimation method, a tool for estimating 3D velocity fields is described which is successfully applied to a living cell sequence. With the estimated velocity fields, motion patterns can be observed, which are a useful starting point for the analysis of dynamic processes in living cells.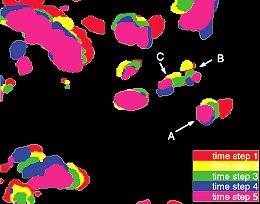
Bibtex Entry
@Article{BergsmaCytometry2001,
author = "Bergsma, C. B. J. and Streekstra, G. J. and Smeulders, A. W. M. and Manders, E. M. M.",
title = "Velocity Estimation of Spots in Three-Dimensional Confocal Image Sequences of Living Cells",
journal = "Cytometry",
number = "4",
volume = "43",
pages = "261--272",
year = "2001",
url = "https://ivi.fnwi.uva.nl/isis/publications/2001/BergsmaCytometry2001",
pdf = "https://ivi.fnwi.uva.nl/isis/publications/2001/BergsmaCytometry2001/BergsmaCytometry2001.pdf",
has_image = 1
}